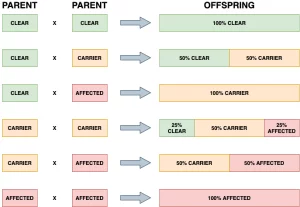
Back in March, CAGT launched a DNA test for progressive retinal atrophy (PRA) in the English Shepherd Dog breed (PRA6). Since then we have been working on a manuscript detailing our research and we are very pleased to announce that it was published last Sunday in Genes.
Back in March, CAGT launched a DNA test for progressive retinal atrophy (PRA) in the English Shepherd Dog breed (PRA6). Since then we have been working on a manuscript detailing our research and we are very pleased to announce that it was published last Sunday in Genes.
Stanbury K, Schofield EC, McLaughlin B, Forman OP, Mellersh CS. Exonic Short Interspersed Nuclear Element Insertion in FAM161A Is Associated with Autosomal Recessive Progressive Retinal Atrophy in the English Shepherd. Genes. 2024; 15(7):952. doi: 10.3390/genes15070952
 We have
We have  Hundreds of Beagles and their owners came together on Sunday 7th July at scenic
Hundreds of Beagles and their owners came together on Sunday 7th July at scenic  On 1st September, two teams will be heading to London to run the
On 1st September, two teams will be heading to London to run the  The Canine Genetics Centre has had something very special to celebrate this month. Our group leader,
The Canine Genetics Centre has had something very special to celebrate this month. Our group leader, 
 Last week five members of the Canine Genetics Centre (CGC) team visited Helsinki, Finland, to attend the
Last week five members of the Canine Genetics Centre (CGC) team visited Helsinki, Finland, to attend the  Thank you to everyone who has donated to the Canine Genetic Centre’s “
Thank you to everyone who has donated to the Canine Genetic Centre’s “